Future: Parallel & Distributed Processing in R for Everyone
Henrik Bengtsson
Why do we parallelize software?
Parallel & distributed processing can be used to:
speed up processing (wall time)
decrease memory footprint (per machine)
avoid data transfers
Comment: I'll focuses on the first two in this talk.
Definition: Future
- A future is an abstraction for a value that will be available later
- The value is the result of an evaluated expression
- The state of a future is unevaluated or evaluated
Friedman & Wise (1976, 1977), Hibbard (1976), Baker & Hewitt (1977)
Definition: Future
- A future is an abstraction for a value that will be available later
- The value is the result of an evaluated expression
- The state of a future is unevaluated or evaluated
v <- expr
Definition: Future
- A future is an abstraction for a value that will be available later
- The value is the result of an evaluated expression
- The state of a future is unevaluated or evaluated
v <- expr
f <- future(expr) v <- value(f)
Example: Sum of 1:50 and 51:100 in parallel
> library(future)> plan(multiprocess)Example: Sum of 1:50 and 51:100 in parallel
> library(future)> plan(multiprocess)> fa <- future( slow_sum( 1:50 ) )>Example: Sum of 1:50 and 51:100 in parallel
> library(future)> plan(multiprocess)> fa <- future( slow_sum( 1:50 ) )> fb <- future( slow_sum(51:100) )>Example: Sum of 1:50 and 51:100 in parallel
> library(future)> plan(multiprocess)> fa <- future( slow_sum( 1:50 ) )> fb <- future( slow_sum(51:100) )> 1:3[1] 1 2 3>Example: Sum of 1:50 and 51:100 in parallel
> library(future)> plan(multiprocess)> fa <- future( slow_sum( 1:50 ) )> fb <- future( slow_sum(51:100) )> 1:3[1] 1 2 3> value(fa)Example: Sum of 1:50 and 51:100 in parallel
> library(future)> plan(multiprocess)> fa <- future( slow_sum( 1:50 ) )> fb <- future( slow_sum(51:100) )> 1:3[1] 1 2 3> value(fa)[1] 1275Example: Sum of 1:50 and 51:100 in parallel
> library(future)> plan(multiprocess)> fa <- future( slow_sum( 1:50 ) )> fb <- future( slow_sum(51:100) )> 1:3[1] 1 2 3> value(fa)[1] 1275> value(fb)[1] 3775Example: Sum of 1:50 and 51:100 in parallel
> library(future)> plan(multiprocess)> fa <- future( slow_sum( 1:50 ) )> fb <- future( slow_sum(51:100) )> 1:3[1] 1 2 3> value(fa)[1] 1275> value(fb)[1] 3775> value(fa) + value(fb)[1] 5050Definition: Future
v <- expr
v %<-% expr
Example: Sum of 1:50 and 51:100 in parallel (implicit API)
> library(future)> plan(multiprocess)> a %<-% slow_sum( 1:50 )> b %<-% slow_sum(51:100)> 1:3[1] 1 2 3> a + b[1] 5050Many ways to resolve futures
plan(sequential)plan(multiprocess)plan(cluster, workers = c("n1", "n2", "n3"))plan(cluster, workers = c("remote1.org", "remote2.org"))...Many ways to resolve futures
plan(sequential)plan(multiprocess)plan(cluster, workers = c("n1", "n2", "n3"))plan(cluster, workers = c("remote1.org", "remote2.org"))...> a %<-% slow_sum( 1:50 )> b %<-% slow_sum(51:100)> a + b[1] 5050R package: future 

- "Write once, run anywhere"
- A simple unified API ("interface of interfaces")
- 100% cross platform
- Easy to install (~0.4 MiB total)
- Very well tested, lots of CPU mileage, production ready
╔════════════════════════════════════════════════════════╗ ║ < Future API > ║ ║ ║ ║ future(), value(), %<-%, ... ║ ╠════════════════════════════════════════════════════════╣ ║ future ║ ╠════════════════════════════════╦═══════════╦═══════════╣ ║ parallel ║ globals ║ (listenv) ║ ╠══════════╦══════════╦══════════╬═══════════╬═══════════╝ ║ snow ║ Rmpi ║ nws ║ codetools ║ ╚══════════╩══════════╩══════════╩═══════════╝
Why a Future API? Problem: heterogeneity
- Different parallel backends ⇔ different APIs
- Choosing API/backend, limits user's options
x <- list(a = 1:50, b = 51:100)y <- lapply(x, FUN = slow_sum)Why a Future API? Problem: heterogeneity
- Different parallel backends ⇔ different APIs
- Choosing API/backend, limits user's options
x <- list(a = 1:50, b = 51:100)y <- lapply(x, FUN = slow_sum)y <- parallel::mclapply(x, FUN = slow_sum)Why a Future API? Problem: heterogeneity
- Different parallel backends ⇔ different APIs
- Choosing API/backend, limits user's options
x <- list(a = 1:50, b = 51:100)y <- lapply(x, FUN = slow_sum)y <- parallel::mclapply(x, FUN = slow_sum)library(parallel)cluster <- makeCluster(4)y <- parLapply(cluster, x, fun = slow_sum)stopCluster(cluster)Why a Future API? Solution: "interface of interfaces"
The Future API encapsulates heterogeneity
- fever decisions for developer to make
- more power to the end user
Philosophy:
- developer decides what to parallelize - user decides how to
Provides atomic building blocks for richer parallel constructs, e.g. 'foreach' and 'future.apply'
Easy to implement new backends, e.g. 'future.batchtools' and 'future.callr'
Why a Future API? 99% Worry Free
- Globals: automatically identified & exported
- Packages: automatically identified & exported
- Static-code inspection by walking the AST
x <- rnorm(n = 100)y <- future({ slow_sum(x) })Why a Future API? 99% Worry Free
- Globals: automatically identified & exported
- Packages: automatically identified & exported
- Static-code inspection by walking the AST
x <- rnorm(n = 100)y <- future({ slow_sum(x) })Globals identified and exported:
slow_sum()- a function (also searched recursively)
Why a Future API? 99% Worry Free
- Globals: automatically identified & exported
- Packages: automatically identified & exported
- Static-code inspection by walking the AST
x <- rnorm(n = 100)y <- future({ slow_sum(x) })Globals identified and exported:
slow_sum()- a function (also searched recursively)x- a numeric vector of length 100
Why a Future API? 99% Worry Free
- Globals: automatically identified & exported
- Packages: automatically identified & exported
- Static-code inspection by walking the AST
x <- rnorm(n = 100)y <- future({ slow_sum(x) })Globals identified and exported:
slow_sum()- a function (also searched recursively)x- a numeric vector of length 100
High Performance Compute (HPC) clusters

Backend: future.batchtools 

- batchtools: Map-Reduce API for HPC schedulers,
e.g. LSF, OpenLava, SGE, Slurm, and TORQUE / PBS - future.batchtools: Future API on top of batchtools
╔═══════════════════════════════════════════════════╗
║ < Future API > ║
╠═══════════════════════════════════════════════════╣
║ future <-> future.batchtools ║
╠═════════════════════════╦═════════════════════════╣
║ parallel ║ batchtools ║
╚═════════════════════════╬═════════════════════════╣
║ SGE, Slurm, TORQUE, ... ║
╚═════════════════════════╝
Real Example: DNA Sequence Analysis
- DNA sequences from 100 cancer patients
- 200 GiB data / patient (~ 10 hours)
raw <- dir(pattern = "[.]fq$")aligned <- listenv()for (i in seq_along(raw)) { aligned[[i]] %<-% DNAseq::align(raw[i])}aligned <- as.list(aligned)Real Example: DNA Sequence Analysis
- DNA sequences from 100 cancer patients
- 200 GiB data / patient (~ 10 hours)
raw <- dir(pattern = "[.]fq$")aligned <- listenv()for (i in seq_along(raw)) { aligned[[i]] %<-% DNAseq::align(raw[i])}aligned <- as.list(aligned)plan(multiprocess)plan(cluster, workers = c("n1", "n2", "n3"))plan(batchtools_sge)
Comment: The use of `listenv` is non-critical and only needed for implicit futures when assigning them by index (instead of by name).
Building on top of Future API

Frontend: future.apply 

- Futurized version of base R's
lapply(),vapply(),replicate(), ... - ... on all future-compatible backends
╔═══════════════════════════════════════════════════════════╗ ║ future_lapply(), future_vapply(), future_replicate(), ... ║ ╠═══════════════════════════════════════════════════════════╣ ║ < Future API > ║ ╠═══════════════════════════════════════════════════════════╣ ║ "wherever" ║ ╚═══════════════════════════════════════════════════════════╝
aligned <- lapply(raw, DNAseq::align)Frontend: future.apply 

- Futurized version of base R's
lapply(),vapply(),replicate(), ... - ... on all future-compatible backends
╔═══════════════════════════════════════════════════════════╗ ║ future_lapply(), future_vapply(), future_replicate(), ... ║ ╠═══════════════════════════════════════════════════════════╣ ║ < Future API > ║ ╠═══════════════════════════════════════════════════════════╣ ║ "wherever" ║ ╚═══════════════════════════════════════════════════════════╝
aligned <- future_lapply(raw, DNAseq::align)Frontend: future.apply 

- Futurized version of base R's
lapply(),vapply(),replicate(), ... - ... on all future-compatible backends
╔═══════════════════════════════════════════════════════════╗ ║ future_lapply(), future_vapply(), future_replicate(), ... ║ ╠═══════════════════════════════════════════════════════════╣ ║ < Future API > ║ ╠═══════════════════════════════════════════════════════════╣ ║ "wherever" ║ ╚═══════════════════════════════════════════════════════════╝
aligned <- future_lapply(raw, DNAseq::align)plan(multiprocess)plan(cluster, workers = c("n1", "n2", "n3"))plan(batchtools_sge)
Frontend: doFuture 

- A foreach adapter on top of the Future API
- Foreach on all future-compatible backends
╔═══════════════════════════════════════════════════════╗
║ foreach API ║
╠════════════╦══════╦════════╦═══════╦══════════════════╣
║ doParallel ║ doMC ║ doSNOW ║ doMPI ║ doFuture ║
╠════════════╩══╦═══╩════════╬═══════╬══════════════════╣
║ parallel ║ snow ║ Rmpi ║ < Future API > ║
╚═══════════════╩════════════╩═══════╬══════════════════╣
║ "wherever" ║
╚══════════════════╝
doFuture::registerDoFuture()plan(batchtools_sge)aligned <- foreach(x = raw) %dopar% { DNAseq::align(x)}1,200+ packages can now parallelize on HPC
┌───────────────────────────────────────────────────────┐
│ │
│ caret, gam, glmnet, plyr, ... (1,200 pkgs) │
│ │
╠═══════════════════════════════════════════════════════╣
║ foreach API ║
╠══════╦════════════╦════════╦═══════╦══════════════════╣
║ doMC ║ doParallel ║ doSNOW ║ doMPI ║ doFuture ║
╠══════╩════════╦═══╩════════╬═══════╬══════════════════╣
║ parallel ║ snow ║ Rmpi ║ < Future API > ║
╚═══════════════╩════════════╩═══════╬══════════════════╣
║ "wherever" ║
╚══════════════════╝
doFuture::registerDoFuture()plan(future.batchtools::batchtools_sge)library(caret)model <- train(y ~ ., data = training)## 2018-05-12> db <- utils::available.packages()> direct <- tools::dependsOnPkgs("foreach", recursive=FALSE, installed=db)> str(direct) chr [1:443] "adabag" "admixturegraph" "ADMMsigma" "Arothron" "ashr" ...> all <- tools::dependsOnPkgs("foreach", recursive=TRUE, installed=db)> str(all) chr [1:1200] "adabag" "admixturegraph" "ADMMsigma" "Arothron" "ashr" ...Futures in the Wild
Frontend: drake - A Workflow Manager 
tasks <- drake_plan( raw_data = readxl::read_xlsx(file_in("raw-data.xlsx")), data = raw_data %>% mutate(Species = forcats::fct_inorder(Species)) %>% select(-X__1), hist = ggplot(data, aes(x = Petal.Width, fill = Species)) + geom_histogram(), fit = lm(Sepal.Width ~ Petal.Width + Species, data), rmarkdown::render(knitr_in("report.Rmd"), output_file = file_out("report.pdf")))future::plan("multiprocess")make(tasks, parallelism = "future")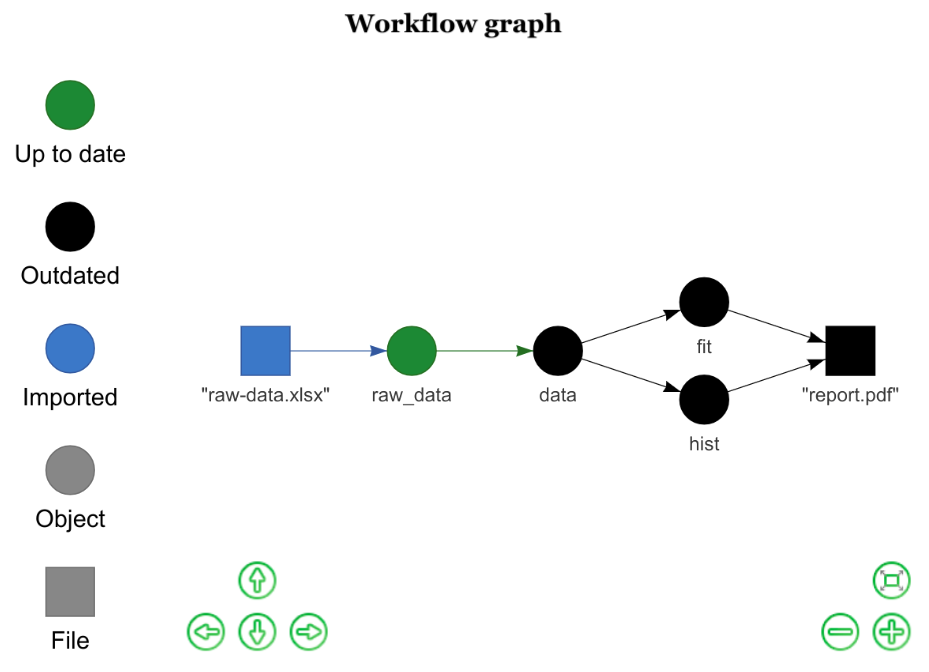
Summary of features
Unified API
Portable code
Worry-free
Developer decides what to parallelize - user decides how to
For beginners as well as advanced users
Nested parallelism on nested heterogeneous backends
Protects against recursive parallelism
Easy to add new backends
Easy to build new frontends
In the near future ...
- Capturing standard output
- Benchmarking (time and memory)
- Killing futures
Building a better future
bug reports,
and suggestions
Thank you!
Appendix (Random Slides)
A1. Features - more details
A1.1 Well Tested
- Large number of unit tests
- System tests
- High code coverage (union of all platform near 100%)
- Cross platform testing
- CI testing
- Testing several R versions (many generations back)
- Reverse package dependency tests
- All backends highly tested
- Large of tests via doFuture across backends on
example():s from foreach, NMF, TSP, glmnet, plyr, caret, etc. (example link)
R Consortium Infrastructure Steering Committee (ISC) Support Project
- Backend Conformance Test Suite - an effort to formalizing and standardizing the above tests into a unified go-to test environment.
A1.2 Nested futures
raw <- dir(pattern = "[.]fq$")aligned <- listenv()for (i in seq_along(raw)) { aligned[[i]] %<-% { chrs <- listenv() for (j in 1:24) { chrs[[j]] %<-% DNAseq::align(raw[i], chr = j) } merge_chromosomes(chrs) }}A1.2 Nested futures
raw <- dir(pattern = "[.]fq$")aligned <- listenv()for (i in seq_along(raw)) { aligned[[i]] %<-% { chrs <- listenv() for (j in 1:24) { chrs[[j]] %<-% DNAseq::align(raw[i], chr = j) } merge_chromosomes(chrs) }}plan(batchtools_sge)
A1.2 Nested futures
raw <- dir(pattern = "[.]fq$")aligned <- listenv()for (i in seq_along(raw)) { aligned[[i]] %<-% { chrs <- listenv() for (j in 1:24) { chrs[[j]] %<-% DNAseq::align(raw[i], chr = j) } merge_chromosomes(chrs) }}plan(batchtools_sge)plan(list(batchtools_sge, sequential))
A1.2 Nested futures
raw <- dir(pattern = "[.]fq$")aligned <- listenv()for (i in seq_along(raw)) { aligned[[i]] %<-% { chrs <- listenv() for (j in 1:24) { chrs[[j]] %<-% DNAseq::align(raw[i], chr = j) } merge_chromosomes(chrs) }}plan(batchtools_sge)plan(list(batchtools_sge, sequential))plan(list(batchtools_sge, multiprocess))
A1.3 Lazy evaluation
By default all futures are resolved using eager evaluation, but the developer has the option to use lazy evaluation.
Explicit API:
f <- future(..., lazy = TRUE)v <- value(f)Implicit API:
v %<-% { ... } %lazy% TRUEA1.4 False-negative & false-positive globals
Identification of globals from static-code inspection has limitations (but defaults cover a large number of use cases):
False negatives, e.g.
my_fcnis not found indo.call("my_fcn", x). Avoid by usingdo.call(my_fcn, x).False positives - non-existing variables, e.g. NSE and variables in formulas. Ignore and leave it to run-time.
x <- "this FP will be exported"data <- data.frame(x = rnorm(1000), y = rnorm(1000))fit %<-% lm(x ~ y, data = data)Comment: ... so, the above works.
A1.5 Full control of globals (explicit API)
Automatic (default):
x <- rnorm(n = 100)y <- future({ slow_sum(x) }, globals = TRUE)By names:
y <- future({ slow_sum(x) }, globals = c("slow_sum", "x"))As name-value pairs:
y <- future({ slow_sum(x) }, globals = list(slow_sum = slow_sum, x = rnorm(n = 100)))Disable:
y <- future({ slow_sum(x) }, globals = FALSE)A1.5 Full control of globals (implicit API)
Automatic (default):
x <- rnorm(n = 100)y %<-% { slow_sum(x) } %globals% TRUEBy names:
y %<-% { slow_sum(x) } %globals% c("slow_sum", "x")As name-value pairs:
y %<-% { slow_sum(x) } %globals% list(slow_sum = slow_sum, x = rnorm(n = 100))Disable:
y %<-% { slow_sum(x) } %globals% FALSEA1.6 Protection: Exporting too large objects
x <- lapply(1:100, FUN = function(i) rnorm(1024 ^ 2))y <- list()for (i in seq_along(x)) { y[[i]] <- future( mean(x[[i]]) )}gives error: "The total size of the 2 globals that need to be exported for the future expression ('mean(x[[i]])') is 800.00 MiB. This exceeds the maximum allowed size of 500.00 MiB (option 'future.globals.maxSize'). There are two globals: 'x' (800.00 MiB of class 'list') and 'i' (48 bytes of class 'numeric')."
A1.6 Protection: Exporting too large objects
x <- lapply(1:100, FUN = function(i) rnorm(1024 ^ 2))y <- list()for (i in seq_along(x)) { y[[i]] <- future( mean(x[[i]]) )}gives error: "The total size of the 2 globals that need to be exported for the future expression ('mean(x[[i]])') is 800.00 MiB. This exceeds the maximum allowed size of 500.00 MiB (option 'future.globals.maxSize'). There are two globals: 'x' (800.00 MiB of class 'list') and 'i' (48 bytes of class 'numeric')."
for (i in seq_along(x)) { x_i <- x[[i]] ## Fix: subset before creating future y[[i]] <- future( mean(x_i) )}Comment: Interesting research project to automate via code inspection.
A1.7 Free futures are resolved
Implicit futures are always resolved:
a %<-% sum(1:10)b %<-% { 2 * a }print(b)## [1] 110A1.7 Free futures are resolved
Implicit futures are always resolved:
a %<-% sum(1:10)b %<-% { 2 * a }print(b)## [1] 110Explicit futures require care by developer:
fa <- future( sum(1:10) )a <- value(fa)fb <- future( 2 * a )A1.7 Free futures are resolved
Implicit futures are always resolved:
a %<-% sum(1:10)b %<-% { 2 * a }print(b)## [1] 110Explicit futures require care by developer:
fa <- future( sum(1:10) )a <- value(fa)fb <- future( 2 * a )For the lazy developer - not recommended (may be expensive):
options(future.globals.resolve = TRUE)fa <- future( sum(1:10) )fb <- future( 2 * value(fa) )A1.8 What's under the hood?
Future class and corresponding methods:
abstract S3 class with common parts implemented,
e.g. globals and protectionnew backends extend this class and implement core methods,
e.g.value()andresolved()built-in classes implement backends on top the parallel package
A1.9 Universal union of parallel frameworks
| future | parallel | foreach | batchtools | BiocParallel | |
|---|---|---|---|---|---|
| Synchronous | ✓ | ✓ | ✓ | ✓ | ✓ |
| Asynchronous | ✓ | ✓ | ✓ | ✓ | ✓ |
| Uniform API | ✓ | ✓ | ✓ | ✓ | |
| Extendable API | ✓ | ✓ | ✓ | ✓ | |
| Globals | ✓ | (✓) | |||
| Packages | ✓ | ||||
| Map-reduce ("lapply") | ✓ | ✓ | foreach() |
✓ | ✓ |
| Load balancing | ✓ | ✓ | ✓ | ✓ | ✓ |
| For loops | ✓ | ||||
| While loops | ✓ | ||||
| Nested config | ✓ | ||||
| Recursive protection | ✓ | mc | mc | mc | mc |
| RNG stream | ✓+ | ✓ | doRNG | (soon) | SNOW |
| Early stopping | ✓ | ✓ | |||
| Traceback | ✓ | ✓ |
A2 Bells & whistles
A2.1 availableCores() & availableWorkers()
availableCores()is a "nicer" version ofparallel::detectCores()that returns the number of cores allotted to the process by acknowledging known settings, e.g.getOption("mc.cores")- HPC environment variables, e.g.
NSLOTS,PBS_NUM_PPN,SLURM_CPUS_PER_TASK, ... _R_CHECK_LIMIT_CORES_
availableWorkers()returns a vector of hostnames based on:- HPC environment information, e.g.
PE_HOSTFILE,PBS_NODEFILE, ... - Fallback to
rep("localhost", availableCores())
- HPC environment information, e.g.
Provide safe defaults to for instance
plan(multiprocess)plan(cluster)A2.2: makeClusterPSOCK()
makeClusterPSOCK():
Improves upon
parallel::makePSOCKcluster()Simplifies cluster setup, especially remote ones
Avoids common issues when workers connect back to master:
- uses SSH reverse tunneling
- no need for port-forwarding / firewall configuration
- no need for DNS lookup
Makes option
-l <user>optional (such that~/.ssh/configis respected)
A2.3 HPC resource parameters
With 'future.batchtools' one can also specify computational resources, e.g. cores per node and memory needs.
plan(batchtools_sge, resources = list(mem = "128gb"))y %<-% { large_memory_method(x) }Specific to scheduler: resources is passed to the job-script template
where the parameters are interpreted and passed to the scheduler.
A2.3 HPC resource parameters
With 'future.batchtools' one can also specify computational resources, e.g. cores per node and memory needs.
plan(batchtools_sge, resources = list(mem = "128gb"))y %<-% { large_memory_method(x) }Specific to scheduler: resources is passed to the job-script template
where the parameters are interpreted and passed to the scheduler.
Each future needs one node with 24 cores and 128 GiB of RAM:
resources = list(l = "nodes=1:ppn=24", mem = "128gb")A3. More Examples
A3.1 Plot remotely - display locally
> library(future)> plan(cluster, workers = "remote.org")A3.1 Plot remotely - display locally
> library(future)> plan(cluster, workers = "remote.org")## Plot remotely> g %<-% R.devices::capturePlot({ filled.contour(volcano, color.palette = terrain.colors) title("volcano data: filled contour map") })A3.1 Plot remotely - display locally
> library(future)> plan(cluster, workers = "remote.org")## Plot remotely> g %<-% R.devices::capturePlot({ filled.contour(volcano, color.palette = terrain.colors) title("volcano data: filled contour map") })## Display locally> gA3.1 Plot remotely - display locally
> library(future)> plan(cluster, workers = "remote.org")## Plot remotely> g %<-% R.devices::capturePlot({ filled.contour(volcano, color.palette = terrain.colors) title("volcano data: filled contour map") })## Display locally> g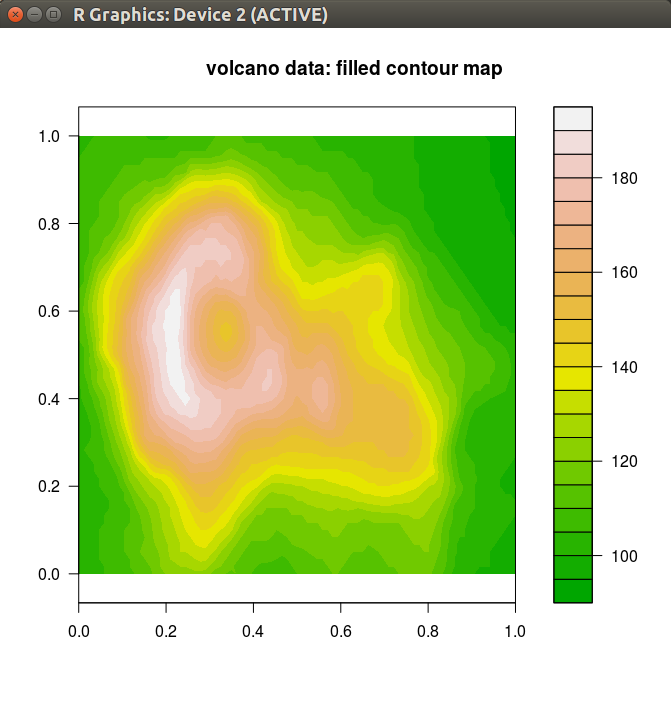
- R (>= 3.3.0)
recordPlot()+replayPlot()- Replotted using local R plot routines
- X11 and similar is not in play here!
A3.2 Profile code remotely - display locally
> plan(cluster, workers = "remote.org")> dat <- data.frame(+ x = rnorm(50e3),+ y = rnorm(50e3)+ )## Profile remotely> p %<-% profvis::profvis({+ plot(x ~ y, data = dat)+ m <- lm(x ~ y, data = dat)+ abline(m, col = "red")+ })A3.2 Profile code remotely - display locally
> plan(cluster, workers = "remote.org")> dat <- data.frame(+ x = rnorm(50e3),+ y = rnorm(50e3)+ )## Profile remotely> p %<-% profvis::profvis({+ plot(x ~ y, data = dat)+ m <- lm(x ~ y, data = dat)+ abline(m, col = "red")+ })## Browse locally> pA3.2 Profile code remotely - display locally
> plan(cluster, workers = "remote.org")> dat <- data.frame(+ x = rnorm(50e3),+ y = rnorm(50e3)+ )## Profile remotely> p %<-% profvis::profvis({+ plot(x ~ y, data = dat)+ m <- lm(x ~ y, data = dat)+ abline(m, col = "red")+ })## Browse locally> p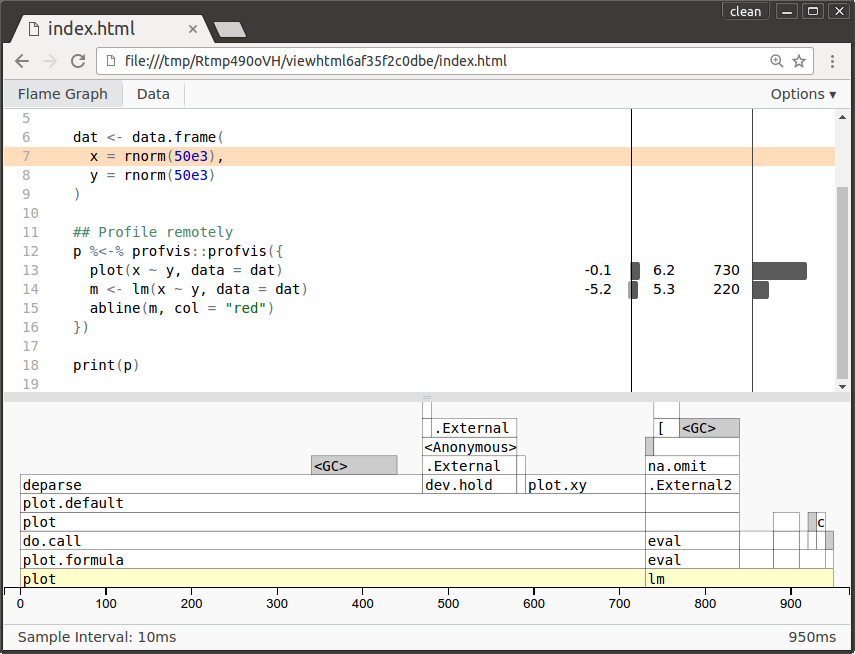
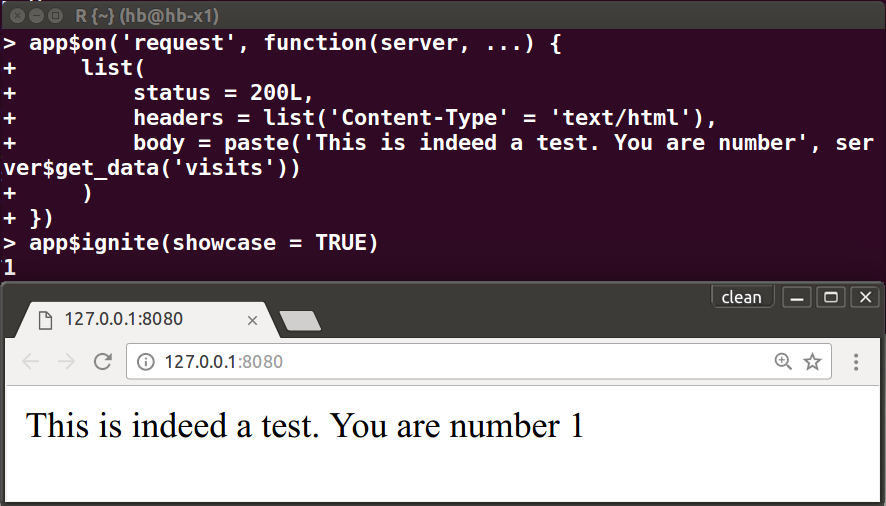
A3.3 fiery - flexible lightweight web server 
"... framework for building web servers in R. ... from serving static
content to full-blown dynamic web-apps"
A3.4 "It kinda just works" (furrr = future + purrr)
plan(multisession)mtcars %>% split(.$cyl) %>% map(~ future(lm(mpg ~ wt, data = .x))) %>% values %>% map(summary) %>% map_dbl("r.squared")## 4 6 8 ## 0.5086326 0.4645102 0.4229655Comment: This approach not do load balancing. I have a few ideas how support for this may be implemented in future framework, which would be beneficial here and elsewhere.
A3.5 Backend: Google Cloud Engine Cluster
library(googleComputeEngineR)vms <- lapply(paste0("node", 1:10), FUN = gce_vm, template = "r-base")cl <- as.cluster(lapply(vms, FUN = gce_ssh_setup), docker_image = "henrikbengtsson/r-base-future")plan(cluster, workers = cl)data <- future_lapply(1:100, montecarlo_pi, B = 10e3)pi_hat <- Reduce(calculate_pi, data)print(pi_hat)## 3.1415A4. Future Work
A4.1 Standard resource types(?)
For any type of futures, the develop may wish to control:
- memory requirements, e.g.
future(..., memory = 8e9) - local machine only, e.g.
remote = FALSE - dependencies, e.g.
dependencies = c("R (>= 3.5.0)", "rio")) - file-system availability, e.g.
mounts = "/share/lab/files" - data locality, e.g.
vars = c("gene_db", "mtcars") - containers, e.g.
container = "docker://rocker/r-base" - generic resources, e.g.
tokens = c("a", "b") - ...?
Risk for bloating the Future API: Which need to be included? Don't want to reinvent the HPC scheduler and Spark.

